Much of the data we collect as ecologists can be plotted as dots on a map. These dots might represent the locations of individual trees or plants, or places where more mobile animals have been spotted. Spatial point pattern data like these are widely used to estimate abundance, map species distributions, and to understand movement behaviour and habitat preferences. Ecology has developed a suite of methods for analysing these point pattern data, often by aggregating the points together into counts, or considering the point locations as presences in presence-absence analyses (with workarounds to conjure up appropriate absence records).
Over roughly the same period, the statistics research community has developed a cohesive framework for considering point data: point process models (PPMs). Spatial PPMs consider the entire pattern of dots as arising from some spatial process. This process might be a continuous ‘intensity surface’ describing how many dots we would expected to see in a given area, or some mechanism by which the points interact with one another to generate the pattern.

From the middle distance, Janine Illian (University of St. Andrews) recaps some of her recent PPM research
In the last few years, papers in the ecology literature have started to draw links to these PPM approaches, and apply them to ecological problems. The popular species distribution modelling (SDM) approach MaxEnt is equivalent to a PPM, and many other SDM approaches are similar to PPMs. Spatial capture recapture and density surface models have been developed which use point processes to estimate abundance from observations of individual locations. Point process models have also been applied to infer habitat preferences from telemetry data.
In June of this year, the Quantitative Ecology SIG sponsored a working group meeting to discuss recent advances in ecological PPMs and identify hurdles to the models being used more widely in ecology. The meeting; Point Processes for Ecology: State of the art and next steps (held in Seattle to coincide with the ISEC 2016 conference) brought together 20 experts in PPMs, analysing ecological point data, and developing statistical software. We wanted to unite people working on PPMs in different corners of ecology, so we deliberately invited researchers who hadn’t previously worked together. Judging by the co-authorship network below, I think we did a decent job of that.
Co-authorship network of workshop attendees – links between people indicated they had previously published papers together. Most of the workshop attendees hadn’t worked together before and were meeting one another for the first time.
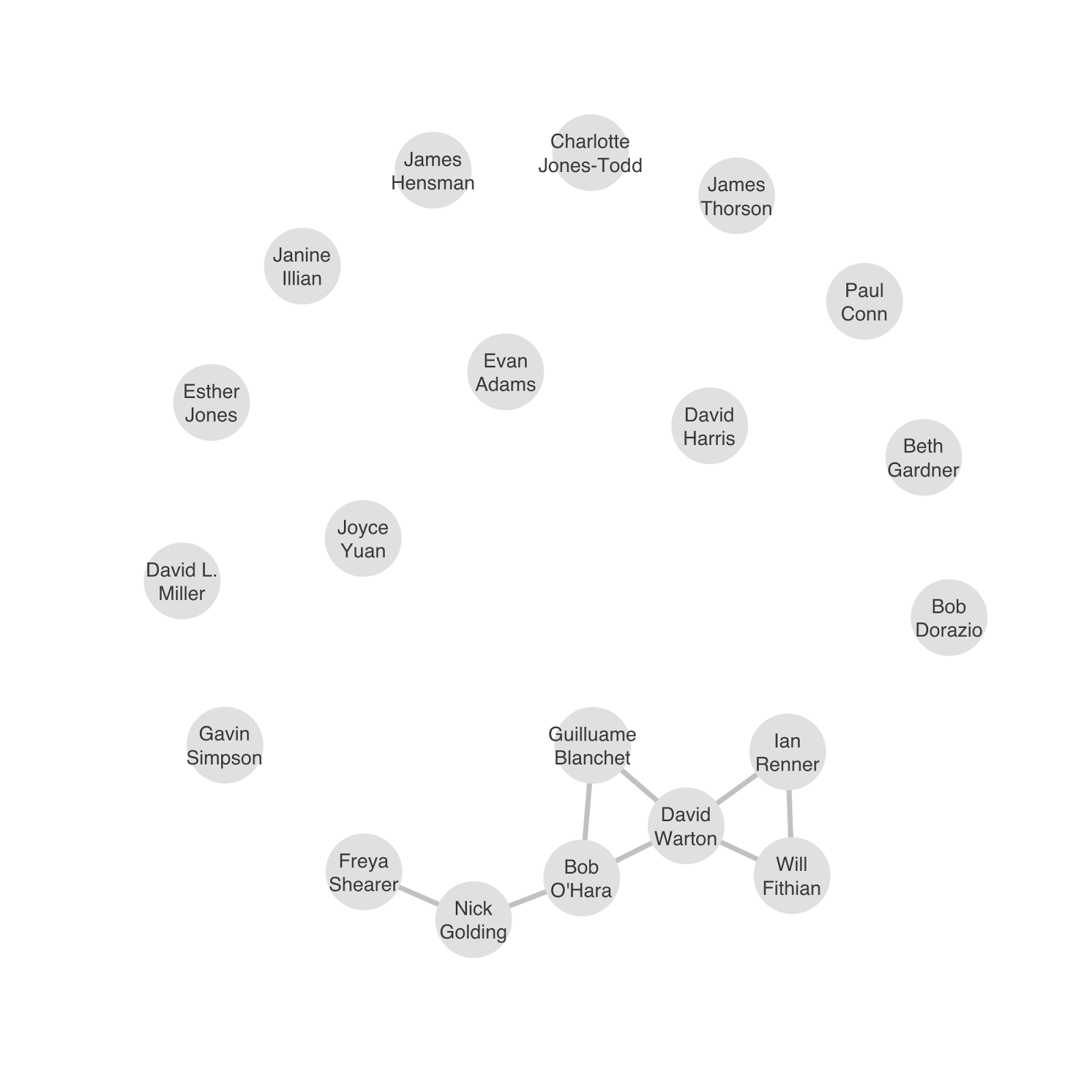
Co-authorship network of workshop attendees. Links between people indicate they had previously published papers together. Most of the workshop attendees hadn’t worked together before and were meeting for the first time.
After kicking off the meeting with the usual introductions and presentations of our own PPM interests, we had an open discussion of what we thought is stopping ecologists from using these advances in their work. This brainstorming session led to a whole lot of messy scribbling on blackboards and, after the chalk dust had settled, we arrived at two broad areas that most needed work to advance ecological PPMs: increasing understanding, and developing tools.
Increasing Understanding
One of the major barriers to uptake of PPMs in ecology is a lack of familiarity with the PPM literature. Despite the high level of statistical literacy in ecology, and ecologists’ wide uptake of similar statistical approaches, most of the ecologists we had spoken with are not at all familiar with PPMs. Getting the message out there with ‘explainer’ papers in the ecological literature is one way of doing that. These sorts of papers have been hugely effective for communicating new methods in the world of species distribution modelling, and a recent paper (by some of the workshop attendees) has done the same to communicate PPM methods to SDM researchers.
We reckon there’s plenty of space for more PPM explainer papers in other areas of ecology, and we settled on a couple of key topics. These include: the benefits of interpreting and explicitly modelling species presence/absence data as arising abundances, rather than some loosely defined probability of presence (a fundamental principle of the PPM approach); ways in which different data types (such as counts, presence/absence surveys and camera trap data) can be combined into a single analysis using PPMs; and the methods that have been developed in the statistical literature for evaluating point pattern models and data.
Developing Tools
Understanding how PPMs work and why they are useful is a critical first step for helping ecologists use these methods. However new methods are useful if ecologists have the tools to apply them. In the case of PPMs, there’s some work to be done making software both available and usable for ecologists. Whilst there is already some great software out there for modelling of point patterns (such as the spatstat and INLA R packages), most of this is targeted at statisticians. Developing software that fits ecologists’ needs, and can be used to answer ecological questions would help us to overcome the technical barriers to PPM uptake.
Specifically, there’s a need to be able to fit PPMs to large, broad-scale ecology datasets efficiently, so a comparison of current and novel methods in that area would be useful. Providing a user-friendly interface to existing and new PPM methods in R (the lingua franca of quantitative ecology) would also be helpful. Linking up a package like that with the ‘explainer’ papers on data integration and model evaluation would ease the route into PPM modelling for ecologists.

Just some of the scribbling on one of many blackboards.
After defining these broad areas for future work, we started fleshing and setting up some of these projects, and even broke some ground writing manuscripts. Hopefully it won’t be long before the co-authorship network becomes a lot more connected. As well as working on these projects, we’re also very keen to hold more events on ecological PPMs in the future; both more working groups and symposia that so that more people can join us. So keep an eye out for more PPM events and papers in the near future!

The nearest we got to a group photo; relaxing in the sun at Gasworks Park, Seattle